UPDATE: A groundbreaking study released on July 5, 2025, reveals new 3D deep learning techniques that could revolutionize fruit tissue analysis. Researchers from KU Leuven, led by Pieter Verboven, have developed a model that outperforms traditional 2D methods, achieving unprecedented accuracy in analyzing the microstructure of apple and pear tissues.
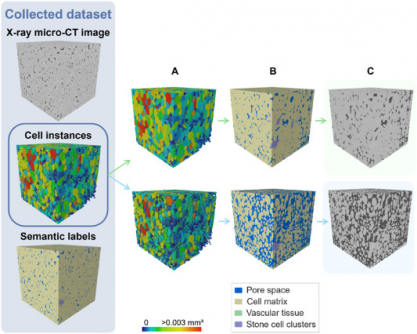
This transformative approach leverages X-ray micro-CT technology to provide non-destructive imaging of plant samples. Traditional microscopy often requires extensive sample preparation and yields limited views, making it difficult to quantify tissue morphology. The new deep learning model addresses these challenges by automating the segmentation of complex plant structures, significantly enhancing the speed and accuracy of research.
The study employed a sophisticated 3D panoptic segmentation framework, achieving an impressive Aggregated Jaccard Index (AJI) of 0.889 for apples and 0.773 for pears. In comparison, previous 2D models scored 0.861 and 0.732, respectively. This leap in performance is crucial for scientists aiming to understand how cellular arrangements impact fruit quality, storability, and susceptibility to disorders like browning and watercore.
Researchers trained the model using augmented datasets that included synthetic data, allowing for comprehensive labeling of fruit tissue microstructures. Critical features such as vascular tissues and stone cell clusters were identified with high precision. Visual validations confirmed the model’s accuracy in detecting vascular bundles in varieties like ‘Kizuri’ and ‘Braeburn’ apples.
As the demand for efficient agricultural practices grows, this study highlights the urgent need for innovative technologies in plant research. The model’s ability to automate tissue characterization can drastically reduce manual labor, allowing scientists to focus on critical analyses. This is particularly significant in fruit research, where understanding microstructures can lead to improvements in crop development and sustainability.
Moreover, the 3D deep learning model is compatible with standard X-ray micro-CT instruments, making it an accessible tool for researchers worldwide. This advancement not only streamlines current methodologies but also opens new avenues for studying tissue development, ripening, and stress responses across various crops.
As this research propels forward, it promises to reshape our approach to plant science, improving how we investigate the intricate relationships between plant anatomy and environmental factors. The implications for agriculture, food science, and ecological sustainability are profound.
Stay tuned for more updates as this story develops. The future of plant research is evolving rapidly, and this breakthrough is just the beginning.