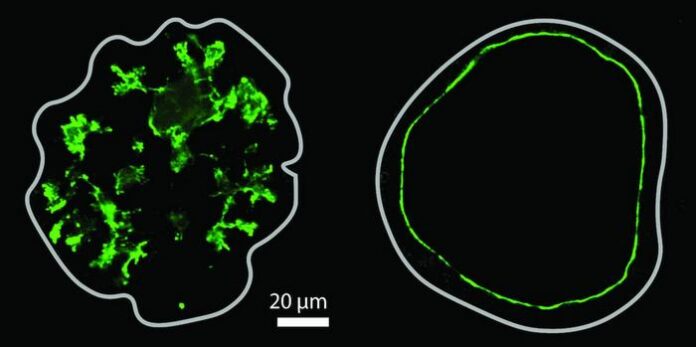
Research led by scientists at the University of Washington has introduced a groundbreaking technique known as Deaminase-Assisted single-molecule chromatin Fiber sequencing (DAF-seq). This innovative approach allows for the detailed mapping of chromatin fiber architectures in single diploid cells, enhancing the understanding of gene regulation.
Gene regulation involves the cooperative binding of proteins along chromatin fibers that stretch across entire chromosomes. Despite its importance, the variation in protein occupancy between different haplotypes and individual cells has not been fully understood in diploid organisms. The DAF-seq technique aims to resolve this issue by enabling near-nucleotide resolution of single-molecule footprinting while simultaneously profiling chromatin states and DNA sequences.
DAF-seq sheds light on the dynamics of protein occupancy at regulatory elements. The method reveals how somatic variants and rare chromatin epialleles affect gene regulation. Notably, single-cell DAF-seq produces comprehensive maps of protein co-occupancy across approximately 99% of the mappable genome in individual cells. This level of detail unveils significant chromatin plasticity, showing that chromatin actuation diverges by 61% between haplotypes within a single cell and by 63% between different cells.
The researchers discovered that regulatory elements tend to co-actuate along the same chromatin fiber in a manner dependent on their distance from one another, closely resembling the behavior of cohesin-mediated loops. This finding suggests a complex regulatory landscape that could inform future studies on gene expression.
The implications of this research extend beyond basic science. Understanding the regulatory mechanisms at play within chromatin could lead to advancements in fields such as medical genetics and cancer research. By characterizing protein occupancy at a single-nucleotide, single-molecule, single-haplotype, and single-cell level, DAF-seq provides a powerful tool for exploring the nuances of gene regulation.
This study was supported by various grants, including those from the National Institutes of Health and the Burroughs Wellcome Fund. The research team, comprising A.B. Stergachis, E.G. Stergachis, and others, acknowledges the contributions of multiple institutions and laboratories that facilitated the study.
The publication of these findings marks a significant advancement in genomic research, promising to enhance our understanding of the intricacies of gene regulation in diploid organisms. As this technology continues to develop, it could pave the way for new insights in genetics and molecular biology.